From pandas to scikit-learn - An Exciting New Workflow
Scikit-Learn’s new integration with Pandas
Scikit-Learn will make one of its biggest upgrades in recent years with its mammoth version 0.20 release. For many data scientists, a typical workflow consists of using Pandas to do exploratory data analysis before moving to scikit-learn for machine learning. This new release will make the process simpler, more feature-rich, robust, and standardized.
Become an Expert
If you want to be trusted to make decisions using pandas and scikit-learn, you must become an expert. I have completely mastered both libraries and have developed special techniques that will massively improve your ability and efficiency to do data analysis and machine learning.
- Master Data Analysis with Python — A comprehensive course with 800+ pages, 500+ exercises, multiple projects, detailed solutions, and video lessons that will help you become a pandas expert.
- Master Machine Learning with Python — My comprehensive guide to become an expert at both the concepts and tools of building a machine learning workflow using Python with scikit-learn.
- Get a sample of the material by enrolling in the FREE Intro to Pandas course.
- Learn directly with me by taking an interactive and fun in-person bootcamp.
Summary and goals of this article
- This article is aimed at those that use Scikit-Learn as their machine learning library but depend on Pandas as their data exploratory and preparation tool.
- It assumes you have some familiarity with both Scikit-Learn and Pandas
- We explore the new
ColumnTransformerestimator, which allows us to apply separate transformations to different subsets of your data in parallel before concatenating the results together. - A major pain point for users (and in my opinion the worst part of Scikit-Learn) was preparing a pandas DataFrame with string values in its columns. This process should become much more standardized.
- The
OneHotEncoderestimator was given a nice upgrade to encode columns with string values. - To help with one hot encoding, we use the new
SimpleImputerestimator to fill in missing values with constants - We will build a custom estimator that does all the “basic” transformations on a DataFrame instead of relying on the built-in Scikit-Learn tools. This will also transform the data with a couple different features not present within Scikit-Learn.
- Finally, we explore binning numeric columns with the new
KBinsDiscretizerestimator.
A note before we get started
This tutorial is provided as a preview of things to come. The final version 0.20 has not been released. It is very likely that this tutorial will be updated at a future date to reflect any changes.
Continuing…
For those that use Pandas as their exploratory and preparation tool before moving to Scikit-Learn for machine learning, you are likely familiar with the non-standard process of handling columns containing string columns. Scikit-Learn’s machine learning models require the input to be a two-dimensional data structure of numeric values. No string values are allowed. Scikit-Learn never provided a canonical way to handle columns of strings, a very common occurrence in data science.
This lead to numerous tutorials all handling string columns in their own way. Some solutions included turning to Pandas get_dummies function. Some used Scikit-Learn’sLabelBinarizer which does one-hot encoding but was designed for labels (the target variable) and not for the input. Others created their own custom estimators. Even entire packages such as sklearn-pandas were built to support this trouble spot. This lack of standardization made for a painful experience for those wanting to build machine learning models with string columns.
Furthermore, there was poor support for making transformations to specific columns and not to the entire dataset. For instance, it’s very common to standardize continuous features but not categorical features. This will now become much easier.
Upgrading to version 0.20
conda update scikit-learn
or pip:
pip install -U scikit-learn
Introducing ColumnTransformer and the upgraded OneHotEncoder
With the upgrade to version 0.20, many workflows from Pandas to Scikit-Learn should start looking similar. The ColumnTransformer estimator applies a transformation to a specific subset of columns of your Pandas DataFrame (or array).
The OneHotEncoder estimator is not new but has been upgraded to encode string columns. Before, it only encoded columns containing numeric categorical data.
Let’s see how these new additions work to handle string columns in a Pandas DataFrame.
Kaggle Housing Dataset
One of Kaggle’s beginning machine learning competitions is the Housing Prices: Advanced Regression Techniques. The goal is to predict housing prices given about 80 features. There is a mix of continuous and categorical columns. You can download the data from the website or use their command line tool (which is very nice).
Inspect the data
Let’s read in our DataFrame and output the first few rows.
>>> import pandas as pd
>>> import numpy as np>>> train = pd.read_csv(‘data/housing/train.csv’)
>>> train.head()
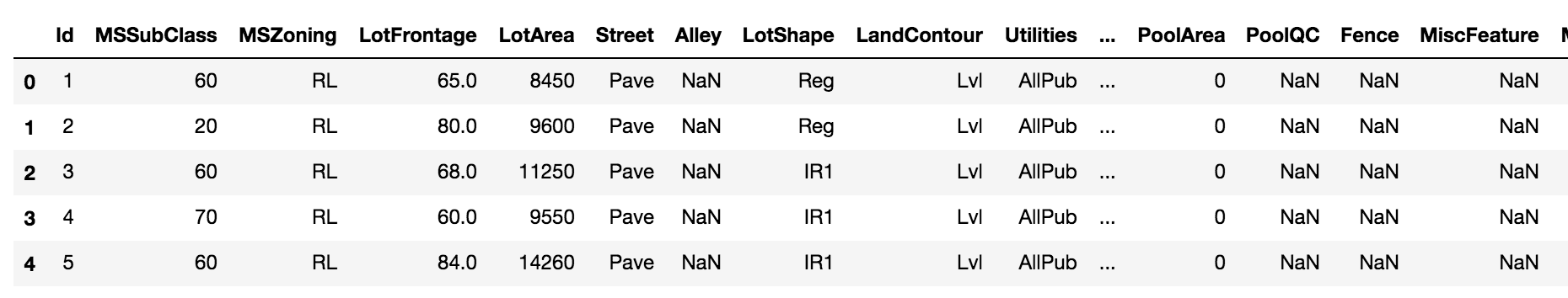
>>> train.shape
(1460, 81)
Remove the target variable from the training set
The target variable is SalePrice which we remove and assign as an array to its own variable. We will use it later when we do machine learning.
>>> y = train.pop('SalePrice').values
Encoding a single string column
To start off, let’s encode a single string column, HouseStyle, which has values for the exterior of the house. Let’s output the unique counts of each string value.
>>> vc = train['HouseStyle'].value_counts()
>>> vc1Story 726
2Story 445
1.5Fin 154
SLvl 65
SFoyer 37
1.5Unf 14
2.5Unf 11
2.5Fin 8
Name: HouseStyle, dtype: int64
We have 8 unique values in this column.
Scikit-Learn Gotcha — Must have 2D data
Most Scikit-Learn estimators require that data be strictly 2-dimensional. If we select the column above as train['HouseStyle'], technically, a Pandas Series is created which is a single dimension of data. We can force Pandas to create a one-column DataFrame, by passing a single-item list to the brackets like this:
>>> hs_train = train[['HouseStyle']].copy()
>>> hs_train.ndim
2
Master Machine Learning with Python
Master Machine Learning with Python is an extremely comprehensive guide that I have written to help you use scikit-learn to do machine learning.
Import, Instantiate, Fit — The three-step process for each estimator
The Scikit-Learn API is consistent for all estimators and uses a three-step process to fit (train) the data.
- Import the estimator we want from the module it’s located in
- Instantiate the estimator, possibly changing its defaults
- Fit the estimator to the data. Possibly transform the data to its new space if need be.
Below, we import OneHotEncoder, instantiate it and ensure that we get a dense (and not sparse) array returned, and then encode our single column with the fit_transform method.
>>> from sklearn.preprocessing import OneHotEncoder
>>> ohe = OneHotEncoder(sparse=False)
>>> hs_train_transformed = ohe.fit_transform(hs_train)
>>> hs_train_transformedarray([[0., 0., 0., ..., 1., 0., 0.],
[0., 0., 1., ..., 0., 0., 0.],
[0., 0., 0., ..., 1., 0., 0.],
...,
[0., 0., 0., ..., 1., 0., 0.],
[0., 0., 1., ..., 0., 0., 0.],
[0., 0., 1., ..., 0., 0., 0.]])
As expected, it has encoded each unique value as its own binary column.
>>> hs_train_transformed.shape(1460, 8)
If you are enjoying this article, consider purchasing the All Access Pass! which includes all my current and future material for one low price.
We have a NumPy array. Where are the column names?
Notice that our output is a NumPy array and not a Pandas DataFrame. Scikit-Learn was not originally built to be directly integrated with Pandas. All Pandas objects are converted to NumPy arrays internally and NumPy arrays are always returned after a transformation.
We can still get our column name from the OneHotEncoder object through its get_feature_names method.
>>> feature_names = ohe.get_feature_names()
>>> feature_namesarray(['x0_1.5Fin', 'x0_1.5Unf', 'x0_1Story', 'x0_2.5Fin',
'x0_2.5Unf', 'x0_2Story', 'x0_SFoyer', 'x0_SLvl'], dtype=object)
Verifying our first row of data is correct
It’s good to verify that our estimator is working properly. Let’s look at the first row of encoded data.
>>> row0 = hs_train_transformed[0]
>>> row0array([0., 0., 0., 0., 0., 1., 0., 0.])
This encodes the 6th value in the array as 1. Let’s use boolean indexing to reveal the feature name.
>>> feature_names[row0 == 1]array(['x0_2Story'], dtype=object)
Now, let’s verify that the first value in our original DataFrame column is the same.
>>> hs_train.values[0]array(['2Story'], dtype=object)
Use inverse_transform to automate this
Just like most transformer objects, there is an inverse_transform method that will get you back your original data. Here we must wrap row0 in a list to make it a 2D array.
>>> ohe.inverse_transform([row0])array([['2Story']], dtype=object)
We can verify all values by inverting the entire transformed array.
>>> hs_inv = ohe.inverse_transform(hs_train_transformed)
>>> hs_invarray([['2Story'],
['1Story'],
['2Story'],
...,
['2Story'],
['1Story'],
['1Story']], dtype=object)>>> np.array_equal(hs_inv, hs_train.values)True
Applying a transformation to the test set
Whatever transformation we do to our training set, we must apply to our test set. Let’s read in the test set and get the same column and apply our transformation.
>>> test = pd.read_csv('data/housing/test.csv')
>>> hs_test = test[['HouseStyle']].copy()
>>> hs_test_transformed = ohe.transform(hs_test)
>>> hs_test_transformedarray([[0., 0., 1., ..., 0., 0., 0.],
[0., 0., 1., ..., 0., 0., 0.],
[0., 0., 0., ..., 1., 0., 0.],
...,
[0., 0., 1., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 1., 0.],
[0., 0., 0., ..., 1., 0., 0.]])
We should again get 8 columns and we do.
>>> hs_test_transformed.shape
(1459, 8)
This example works nicely, but there are multiple cases where we will run into problems. Let’s examine them now.
Trouble area #1 — Categories unique to the test set
What happens if we have a home with a house style that is unique to just the test set? Say something like 3Story. Let's change the first value of the house styles and see what the default is from Scikit-Learn.
>>> hs_test = test[['HouseStyle']].copy()
>>> hs_test.iloc[0, 0] = '3Story'
>>> hs_test.head(3)HouseStyle
0 3Story
1 1Story
2 2Story>>> ohe.transform(hs_test)ValueError: Found unknown categories ['3Story'] in column 0 during transform
Error: Unknown Category
By default, our encoder will produce an error. This is likely what we want as we need to know if there are unique strings in the test set. If you do have this problem then there could be something much deeper that needs investigating. For now, we will ignore the problem and encode this row as all 0’s by setting the handle_unknown parameter to 'ignore' upon instantiation.
>>> ohe = OneHotEncoder(sparse=False, handle_unknown='ignore')
>>> ohe.fit(hs_train)>>> hs_test_transformed = ohe.transform(hs_test)
>>> hs_test_transformedarray([[0., 0., 0., ..., 0., 0., 0.],
[0., 0., 1., ..., 0., 0., 0.],
[0., 0., 0., ..., 1., 0., 0.],
...,
[0., 0., 1., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 1., 0.],
[0., 0., 0., ..., 1., 0., 0.]])
Let’s verify that the first row is all 0's.
>>> hs_test_transformed[0]array([0., 0., 0., 0., 0., 0., 0., 0.])
Trouble area #2 — Missing Values in test set
If you have missing values in your test set (NaN or None), then these will be ignored as long as handle_unknown is set to 'ignore'. Let’s put some missing values in the first couple elements of our test set.
>>> hs_test = test[['HouseStyle']].copy()
>>> hs_test.iloc[0, 0] = np.nan
>>> hs_test.iloc[1, 0] = None
>>> hs_test.head(4)HouseStyle
0 NaN
1 None
2 2Story
3 2Story>>> hs_test_transformed = ohe.transform(hs_test)
>>> hs_test_transformed[:4]array([[0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0., 1., 0., 0.],
[0., 0., 0., 0., 0., 1., 0., 0.]])
Trouble area #3 — Missing Values in training set
Missing values in the training set are more of an issue. As of now, the OneHotEncoder estimator cannot fit with missing values.
>>> hs_train = hs_train.copy()
>>> hs_train.iloc[0, 0] = np.nan
>>> ohe = OneHotEncoder(sparse=False, handle_unknown='ignore')
>>> ohe.fit_transform(hs_train)TypeError: '<' not supported between instances of 'str' and 'float'
It would be nice if there was an option to ignore them like what happens when transforming the test set above. As of now, this doesn’t exist and we must impute it.
Must impute missing values
For now, we must impute the missing values. The old Imputer from the preprocessing module got deprecated. A new module, impute, was formed in its place, with a new estimator SimpleImputer and a new strategy, 'constant'. By default, using this strategy will fill missing values with the string ‘missing_value’. We can choose what to set it with the fill_value parameter.
>>> hs_train = train[['HouseStyle']].copy()
>>> hs_train.iloc[0, 0] = np.nan>>> from sklearn.impute import SimpleImputer
>>> si = SimpleImputer(strategy='constant', fill_value='MISSING')
>>> hs_train_imputed = si.fit_transform(hs_train)
>>> hs_train_imputedarray([['MISSING'],
['1Story'],
['2Story'],
...,
['2Story'],
['1Story'],
['1Story']], dtype=object)
From here we can encode as we did previously.
>>> hs_train_transformed = ohe.fit_transform(hs_train_imputed)
>>> hs_train_transformedarray([[0., 0., 0., ..., 1., 0., 0.],
[0., 0., 1., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 0., 0.],
...,
[0., 0., 0., ..., 0., 0., 0.],
[0., 0., 1., ..., 0., 0., 0.],
[0., 0., 1., ..., 0., 0., 0.]])
Notice, that we now have an extra column and and an extra feature name.
>>> hs_train_transformed.shape(1460, 9)>>> ohe.get_feature_names()array(['x0_1.5Fin', 'x0_1.5Unf', 'x0_1Story', 'x0_2.5Fin',
'x0_2.5Unf', 'x0_2Story', 'x0_MISSING', 'x0_SFoyer',
'x0_SLvl'], dtype=object)
More on fit_transform
For all estimators, the fit_transform method will first call the fit method and then call the transform method. The fit method finds the key properties that will be used during the transformation. For instance, with the SimpleImputer, if the strategy were ‘mean’, then it would find the mean of each column during the fit method. It would store this mean for every column. When transform is called, it uses this stored mean of every column to fill in the missing values and returns the transformed array.
The OneHotEncoder works analogously. During the fit method, it finds all the unique values for each column and again stores this. When transform is called, it uses these stored unique values to produce the binary array.
Apply both transformations to the test set
We can manually apply each of the two steps above in order like this:
>>> hs_test = test[['HouseStyle']].copy()
>>> hs_test.iloc[0, 0] = 'unique value to test set'
>>> hs_test.iloc[1, 0] = np.nan>>> hs_test_imputed = si.transform(hs_test)
>>> hs_test_transformed = ohe.transform(hs_test_imputed)
>>> hs_test_transformed.shape(1459, 8)>>> ohe.get_feature_names()array(['x0_1.5Fin', 'x0_1.5Unf', 'x0_1Story', 'x0_2.5Fin',
'x0_2.5Unf', 'x0_2Story', 'x0_SFoyer', 'x0_SLvl'],
dtype=object)
Use a Pipeline instead
Scikit-Learn provides a Pipeline estimator that takes a list of transformations and applies them in succession. You can also run a machine learning model as the final estimator. Here we simply impute and encode.
>>> from sklearn.pipeline import Pipeline
Each step is a two-item tuple consisting of a string that labels the step and the instantiated estimator. The output of the previous step is the input to the next step.
>>> si_step = ('si', SimpleImputer(strategy='constant',
fill_value='MISSING'))
>>> ohe_step = ('ohe', OneHotEncoder(sparse=False,
handle_unknown='ignore'))
>>> steps = [si_step, ohe_step]
>>> pipe = Pipeline(steps)>>> hs_train = train[['HouseStyle']].copy()
>>> hs_train.iloc[0, 0] = np.nan
>>> hs_transformed = pipe.fit_transform(hs_train)
>>> hs_transformed.shape(1460, 9)
The test set is easily transformed through each step of the pipeline by simply passing it to the transform method.
>>> hs_test = test[['HouseStyle']].copy()
>>> hs_test_transformed = pipe.transform(hs_test)
>>> hs_test_transformed.shape(1459, 9)
Why just the transform method for the test set?
When transforming the test set, it's important to just call the transform method and not fit_transform. When we ran fit_transform on the training set, Scikit-Learn found all the necessary information it needed in order to transform any other dataset containing the same column names.
Transforming Multiple String Columns
Encoding multiple string columns is not a problem. Select the columns you want and then pass the new DataFrame through the same pipeline again.
>>> string_cols = ['RoofMatl', 'HouseStyle']
>>> string_train = train[string_cols]
>>> string_train.head(3)RoofMatl HouseStyle
0 CompShg 2Story
1 CompShg 1Story
2 CompShg 2Story>>> string_train_transformed = pipe.fit_transform(string_train)
>>> string_train_transformed.shape(1460, 16)
Get individual pieces of the pipeline
It is possible to retrieve each individual transformer within the pipeline through its name from the named_steps dictionary atribute. In this instance, we get the one-hot encoder so that we can output the feature names.
>>> ohe = pipe.named_steps['ohe']
>>> ohe.get_feature_names()array(['x0_ClyTile', 'x0_CompShg', 'x0_Membran', 'x0_Metal',
'x0_Roll', 'x0_Tar&Grv', 'x0_WdShake', 'x0_WdShngl',
'x1_1.5Fin', 'x1_1.5Unf', 'x1_1Story', 'x1_2.5Fin',
'x1_2.5Unf', 'x1_2Story', 'x1_SFoyer', 'x1_SLvl'],
dtype=object)
Use the new ColumnTransformer to choose columns
The brand new ColumnTransformer (part of the new compose module) allows you to choose which columns get which transformations. Categorical columns will almost always need separate transformations than continuous columns.
The ColumnTransformer is currently experimental, meaning that its functionality can change in the future.
The ColumnTransformer takes a list of three-item tuples. The first value in the tuple is a name that labels it, the second is an instantiated estimator, and the third is a list of columns you want to apply the transformation to. The tuple will look like this:
('name', SomeTransformer(parameters), columns)
The columns actually don’t have to be column names. Instead, you can use the integer indexes of the columns, a boolean array, or even a function (which accepts the entire DataFrame as the argument and must return a selection of columns).
You can also use NumPy arrays with the ColumnTransformer, but this tutorial is focused on the integration of Pandas so we will stick with just using DataFrames.
Master Python, Data Science and Machine Learning
Immerse yourself in my comprehensive path for mastering data science and machine learning with Python. Purchase the All Access Pass to get lifetime access to all current and future courses. Some of the courses it contains:
- Master the Fundamentals of Python— A comprehensive introduction to Python (300+ pages, 150+ exercises)
- Master Data Analysis with Python — The most comprehensive course available to learn pandas. (800+ pages and 500+ exercises)
- Master Machine Learning with Python — A deep dive into doing machine learning with scikit-learn constantly updated to showcase the latest and greatest tools. (300+ pages)
Pass a Pipeline to the ColumnTransformer
We can even pass a pipeline of many transformations to the column transformer, which is what we do here because we have multiple transformations on our string columns.
Below, we reproduce the above imputation and encoding using the ColumnTransformer. Notice that the pipeline is the exact same as above, just with cat appended to each variable name. We will add a different pipeline for the numeric columns in an upcoming section.
>>> from sklearn.compose import ColumnTransformer>>> cat_si_step = ('si', SimpleImputer(strategy='constant',
fill_value='MISSING'))
>>> cat_ohe_step = ('ohe', OneHotEncoder(sparse=False,
handle_unknown='ignore'))>>> cat_steps = [cat_si_step, cat_ohe_step]
>>> cat_pipe = Pipeline(cat_steps)
>>> cat_cols = ['RoofMatl', 'HouseStyle']
>>> cat_transformers = [('cat', cat_pipe, cat_cols)]
>>> ct = ColumnTransformer(transformers=cat_transformers)
Pass the entire DataFrame to the ColumnTransformer
The ColumnTransformer instance selects the columns we want to use, so we simply pass the entire DataFrame to the fit_transform method. The desired columns will be selected for us.
>>> X_cat_transformed = ct.fit_transform(train)
>>> X_cat_transformed.shape(1460, 16)
We can now transform our test set in the same manner.
>>> X_cat_transformed_test = ct.transform(test)
>>> X_cat_transformed_test.shape(1459, 16)
Retrieving the feature names
We have to do a little digging to get the feature names. All the transformers are stored in the named_transformers_ dictionary attribute. We then use the names, the first item from the three-item tuple to select the specific transformer. Below, we select our transformer (there is only one here — a pipeline named ‘cat’).
>>> pl = ct.named_transformers_['cat']
Then from this pipeline we select the one-hot encoder object and finally get the feature names.
>>> ohe = pl.named_steps['ohe']
>>> ohe.get_feature_names()array(['x0_ClyTile', 'x0_CompShg', 'x0_Membran', 'x0_Metal',
'x0_Roll','x0_Tar&Grv', 'x0_WdShake', 'x0_WdShngl',
'x1_1.5Fin', 'x1_1.5Unf', 'x1_1Story', 'x1_2.5Fin',
'x1_2.5Unf', 'x1_2Story', 'x1_SFoyer', 'x1_SLvl'],
dtype=object)
Transforming the numeric columns
The numeric columns will need a different set of transformations. Instead of imputing missing values with a constant, the median or mean is often chosen. And instead of encoding the values, we usually standardize them by subtracting the mean of each column and dividing by the standard deviation. This helps many models like ridge regression produce a better fit.
Using all the numeric columns
Instead of selecting just one or two columns by hand as we did above with the string columns, we can select all of the numeric columns. We do this by first finding the data type of each column with the dtypes attribute and then testing whether the kind of each dtype is 'O'. The dtypes attribute returns a Series of NumPy dtype objects. Each of these has a kind attribute that is a single character. We can use this to find the numeric or string columns. Pandas stores all of its string columns as object which have a kind equal to ‘O’. See the NumPy docs for more on the kind attribute.
>>> train.dtypes.head()Id int64
MSSubClass int64
MSZoning object
LotFrontage float64
LotArea int64
dtype: object
Get the kinds, a one character string representing the dtype.
>>> kinds = np.array([dt.kind for dt in train.dtypes])
>>> kinds[:5]array(['i', 'i', 'O', 'f', 'i'], dtype='<U1')
Assume all numeric columns are non-object. We can also get the categorical columns in this manner.
>>> all_columns = train.columns.values
>>> is_num = kinds != 'O'
>>> num_cols = all_columns[is_num]
>>> num_cols[:5]array(['Id', 'MSSubClass', 'LotFrontage', 'LotArea', 'OverallQual'],
dtype=object)>>> cat_cols = all_columns[~is_num]
>>> cat_cols[:5]array(['MSZoning', 'Street', 'Alley', 'LotShape', 'LandContour'],
dtype=object)
Once we have our numeric column names, we can use the ColumnTransformer again.
>>> from sklearn.preprocessing import StandardScaler>>> num_si_step = ('si', SimpleImputer(strategy='median'))
>>> num_ss_step = ('ss', StandardScaler())
>>> num_steps = [num_si_step, num_ss_step]>>> num_pipe = Pipeline(num_steps)
>>> num_transformers = [('num', num_pipe, num_cols)]>>> ct = ColumnTransformer(transformers=num_transformers)
>>> X_num_transformed = ct.fit_transform(train)
>>> X_num_transformed.shape(1460, 37)
Combining both categorical and numerical column transformations
We can apply separate transformations to each section of our DataFrame with ColumnTransformer. We will use every single column in this example.
We then create a separate pipeline for both categorical and numerical columns and then use the ColumnTransformer to independently transform them. These two transformations happen in parallel. The results of each are then concatenated together.
>>> transformers = [('cat', cat_pipe, cat_cols),
('num', num_pipe, num_cols)]
>>> ct = ColumnTransformer(transformers=transformers)
>>> X = ct.fit_transform(train)
>>> X.shape(1460, 305)
Machine Learning
The whole point of this exercise is to set up our data so that we can do machine learning. We can create one final pipeline and add a machine learning model as the final estimator. The first step in the pipeline will be the entire transformation we just did above. We assigned y way back at the top of the tutorial as the SalePrice. Here, we will just use thefit method instead of fit_transform since our final step is a machine learning model and does no transformations.
>>> from sklearn.linear_model import Ridge>>> ml_pipe = Pipeline([('transform', ct), ('ridge', Ridge())])
>>> ml_pipe.fit(train, y)
We can evaluate our model with the score method, which returns the R-squared value:
>>> ml_pipe.score(train, y)0.92205
Cross-Validation
Of course, scoring ourselves on the training set is not useful. Let’s do some K-fold cross-validation to get an idea of how well we would do with unseen data. We set a random state so that the splits will be the same throughout the rest of the tutorial.
>>> from sklearn.model_selection import KFold, cross_val_score
>>> kf = KFold(n_splits=5, shuffle=True, random_state=123)
>>> cross_val_score(ml_pipe, train, y, cv=kf).mean()0.813
Selecting parameters when Grid Searching
Grid searching in Scikit-Learn requires us to pass a dictionary of parameter names mapped to possible values. When using a pipeline, we must use the name of the step followed by a double-underscore and then the parameter name. If there are multiple layers to your pipeline, as we have here, we must continue using double-underscores to move up a level until we reach the estimator whose parameters we would like to optimize.
>>> from sklearn.model_selection import GridSearchCV>>> param_grid = {
'transform__num__si__strategy': ['mean', 'median'],
'ridge__alpha': [.001, 0.1, 1.0, 5, 10, 50, 100, 1000],
}
>>> gs = GridSearchCV(ml_pipe, param_grid, cv=kf)
>>> gs.fit(train, y)
>>> gs.best_params_{'ridge__alpha': 10, 'transform__num__si__strategy': 'median'}>>> gs.best_score_0.819
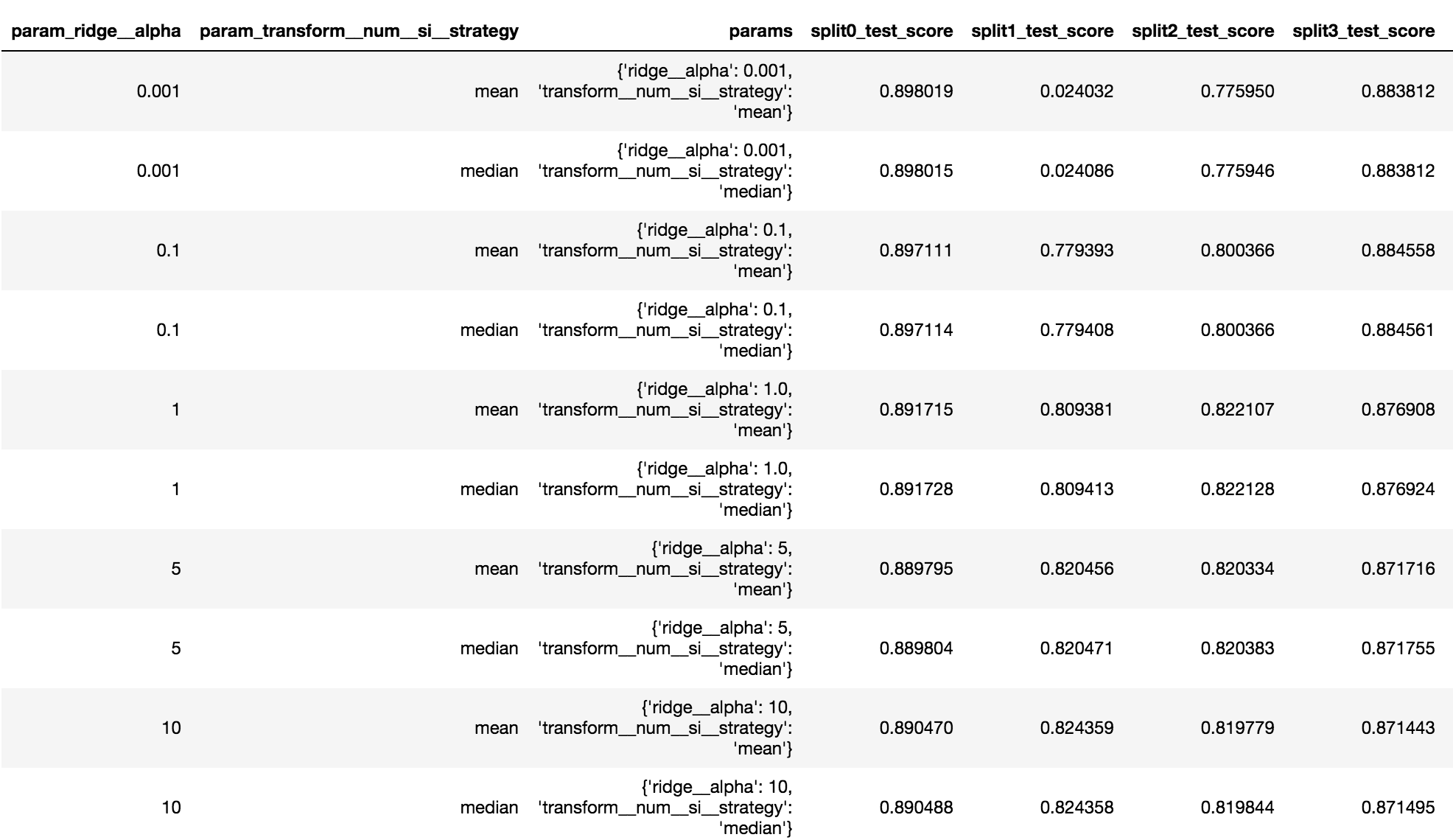
Getting all the grid search results in a Pandas DataFrame
All the results of the grid search are stored in the cv_results_ attribute. This is a dictionary that can get converted to a Pandas DataFrame for a nice display and it provides a structure that is much easier to manually scan.
>>> pd.DataFrame(gs.cv_results_)
Building a custom transformer that does all the basics
There are a few limitations to the above workflow. For instance, it would be nice if the OneHotEncoder gave you the option of ignoring missing values during the fit method. It could simply encode missing values as a row of all zeros. Currently, it forces us to fill the missing values with some string and then encodes this string as a separate column.
Low-frequency strings
Also, string columns that appear only a few times during the training set may not be reliable predictors in the test set. We may want to encode those as if they were missing as well.
Writing your own estimator class
Scikit-Learn provides some help within its documentation on writing your own estimator class. The BaseEstimator class found within the base module provides the get_params and set_params methods for you. The set_params method is necessary when doing a grid search. You can write your own or inherit from the BaseEstimator. There is also a TransformerMixin but it just writes the fit_transform method for you. We do this in one line of code below, so we don’t inherit from it.
The following class BasicTransformer does the following:
- Fills in missing values with either the mean or median for numeric columns
- Standardizes all numeric columns
- Uses one hot encoding for string columns
- Does not fill in missing values for categorical columns. Instead, it encodes them as a 0's
- Ignores unique values in string columns in the test set
- Allows you to choose a threshold for the number of occurrences a value must have in a string column. Strings below this threshold will be encoded as all 0's
- It only works with DataFrames and is just experimental and not tested so it will break for some datasets
- It is called ‘basic’ because, these are probably the most basic transformations that typically get done to many datasets.
from sklearn.base import BaseEstimatorclass BasicTransformer(BaseEstimator):
def __init__(self, cat_threshold=None, num_strategy='median',
return_df=False):
# store parameters as public attributes
self.cat_threshold = cat_threshold
if num_strategy not in ['mean', 'median']:
raise ValueError('num_strategy must be either "mean" or
"median"')
self.num_strategy = num_strategy
self.return_df = return_df
def fit(self, X, y=None):
# Assumes X is a DataFrame
self._columns = X.columns.values
# Split data into categorical and numeric
self._dtypes = X.dtypes.values
self._kinds = np.array([dt.kind for dt in X.dtypes])
self._column_dtypes = {}
is_cat = self._kinds == 'O'
self._column_dtypes['cat'] = self._columns[is_cat]
self._column_dtypes['num'] = self._columns[~is_cat]
self._feature_names = self._column_dtypes['num']
# Create a dictionary mapping categorical column to unique
# values above threshold
self._cat_cols = {}
for col in self._column_dtypes['cat']:
vc = X[col].value_counts()
if self.cat_threshold is not None:
vc = vc[vc > self.cat_threshold]
vals = vc.index.values
self._cat_cols[col] = vals
self._feature_names = np.append(self._feature_names, col
+ '_' + vals)
# get total number of new categorical columns
self._total_cat_cols = sum([len(v) for col, v in
self._cat_cols.items()])
# get mean or median
num_cols = self._column_dtypes['num']
self._num_fill = X[num_cols].agg(self.num_strategy)
return self
def transform(self, X):
# check that we have a DataFrame with same column names as
# the one we fit
if set(self._columns) != set(X.columns):
raise ValueError('Passed DataFrame has different columns
than fit DataFrame')
elif len(self._columns) != len(X.columns):
raise ValueError('Passed DataFrame has different number
of columns than fit DataFrame')
# fill missing values
num_cols = self._column_dtypes['num']
X_num = X[num_cols].fillna(self._num_fill)
# Standardize numerics
std = X_num.std()
X_num = (X_num - X_num.mean()) / std
zero_std = np.where(std == 0)[0]
# If there is 0 standard deviation, then all values are the
# same. Set them to 0.
if len(zero_std) > 0:
X_num.iloc[:, zero_std] = 0
X_num = X_num.values
# create separate array for new encoded categoricals
X_cat = np.empty((len(X), self._total_cat_cols),
dtype='int')
i = 0
for col in self._column_dtypes['cat']:
vals = self._cat_cols[col]
for val in vals:
X_cat[:, i] = X[col] == val
i += 1
# concatenate transformed numeric and categorical arrays
data = np.column_stack((X_num, X_cat))
# return either a DataFrame or an array
if self.return_df:
return pd.DataFrame(data=data,
columns=self._feature_names)
else:
return data
def fit_transform(self, X, y=None):
return self.fit(X).transform(X)
def get_feature_names():
return self._feature_names
Using our BasicTransformer
Our BasicTransformer estimator should be able to be used just like any other scikit-learn estimator. We can instantiate it and then transform our data.
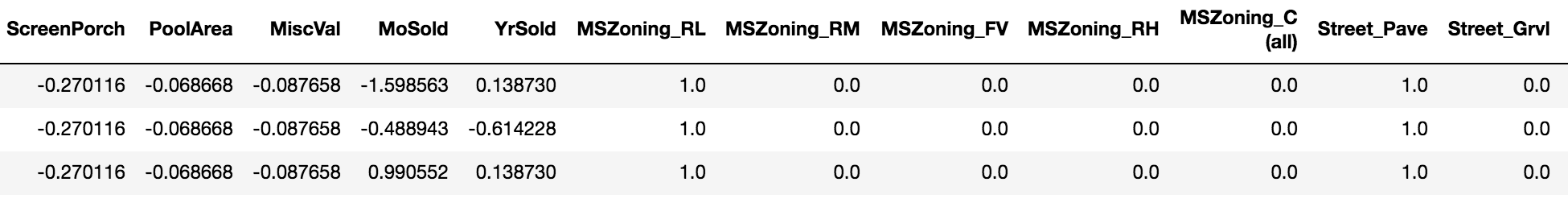
>>> bt = BasicTransformer(cat_threshold=3, return_df=True)
>>> train_transformed = bt.fit_transform(train)
>>> train_transformed.head(3)
Using our transformer in a pipeline
Our transformer can be part of a pipeline.
>>> basic_pipe = Pipeline([('bt', bt), ('ridge', Ridge())])
>>> basic_pipe.fit(train, y)
>>> basic_pipe.score(train, y)0.904
We can also cross-validate with it as well and get a similar score as we did with our scikit-learn column transformer pipeline from above.
>>> cross_val_score(basic_pipe, train, y, cv=kf).mean()0.816
We can use it as part of a grid search as well. It turns out that not including low-count strings did not help this particular model, though it stands to reason it could in other models. The best score did improve a bit, perhaps due to using a slightly different encoding scheme.
>>> param_grid = {
'bt__cat_threshold': [0, 1, 2, 3, 5],
'ridge__alpha': [.1, 1, 10, 100]
}>>> gs = GridSearchCV(p, param_grid, cv=kf)
>>> gs.fit(train, y)
>>> gs.best_params_{'bt__cat_threshold': 0, 'ridge__alpha': 10}>>> gs.best_score_
0.830
Binning and encoding numeric columns with the new KBinsDiscretizer
There are a few columns that contain years. It makes more sense to bin the values in these columns and treat them as categories. Scikit-Learn introduced the new estimator KBinsDiscretizer to do just this. It not only bins the values, but it encodes them as well. Before you could have done this manually with Pandas cut or qcut functions.
Let’s see how it works with just the YearBuilt column.
>>> from sklearn.preprocessing import KBinsDiscretizer
>>> kbd = KBinsDiscretizer(encode='onehot-dense')
>>> year_built_transformed = kbd.fit_transform(train[['YearBuilt']])
>>> year_built_transformedarray([[0., 0., 0., 0., 1.],
[0., 0., 1., 0., 0.],
[0., 0., 0., 1., 0.],
...,
[1., 0., 0., 0., 0.],
[0., 1., 0., 0., 0.],
[0., 0., 1., 0., 0.]])
By default, each bin contains (approximately) an equal number of observations. Let’s sum up each column to verify this.
>>> year_built_transformed.sum(axis=0)array([292., 274., 307., 266., 321.])
This is the ‘quantile’ strategy. You can choose ‘uniform’ to make the bin edges equally spaced or ‘kmeans’ which uses K-means clustering to find the bin edges.
>>> kbd.bin_edges_array([array([1872. , 1947.8, 1965. , 1984. , 2003. , 2010. ])],
dtype=object)
Processing all the year columns separately with ColumnTransformer
We now have another subset of columns that need separate processing and we can do this with the ColumnTransformer. The following code adds one more step to our previous transformation. We also drop the Id column which was just identifying each row.
>>> year_cols = ['YearBuilt', 'YearRemodAdd', 'GarageYrBlt',
'YrSold']
>>> not_year = ~np.isin(num_cols, year_cols + ['Id'])
>>> num_cols2 = num_cols[not_year]>>> year_si_step = ('si', SimpleImputer(strategy='median'))
>>> year_kbd_step = ('kbd', KBinsDiscretizer(n_bins=5,
encode='onehot-dense'))
>>> year_steps = [year_si_step, year_kbd_step]
>>> year_pipe = Pipeline(year_steps)>>> transformers = [('cat', cat_pipe, cat_cols),
('num', num_pipe, num_cols2),
('year', year_pipe, year_cols)]>>> ct = ColumnTransformer(transformers=transformers)
>>> X = ct.fit_transform(train)
>>> X.shape(1460, 320)
We cross-validate and score and see that all this work yielded us no improvements.
>>> ml_pipe = Pipeline([('transform', ct), ('ridge', Ridge())])
>>> cross_val_score(ml_pipe, train, y, cv=kf).mean()
0.813
Using a different number of bins for each column might improve our results. Still, the KBinsDiscretizer makes it easy to bin numeric variables.
More goodies in Scikit-Learn 0.20
There are more new features that come with the upcoming release. Check the What’s New section of the docs for more. There are a ton of changes.
Conclusion
This article introduced a new workflow that will be available to Scikit-Learn users who rely on Pandas for the initial data exploration and preparation. A much smoother and feature-rich process for taking a Pandas DataFrame and transforming it so that it is ready for machine learning is now done through the new and improved estimatorsColumnTransformer, SimpleImputer, OneHotEncoder, and KBinsDiscretizer.
I am very excited to see this new upgrade and am going to be integrating these new workflows immediately into my projects and teaching materials.
Master Python, Data Science and Machine Learning
Immerse yourself in my comprehensive path for mastering data science and machine learning with Python. Purchase the All Access Pass to get lifetime access to all current and future courses. Some of the courses it contains:
- Master the Fundamentals of Python— A comprehensive introduction to Python (300+ pages, 150+ exercises)
- Master Data Analysis with Python — The most comprehensive course available to learn pandas. (800+ pages and 500+ exercises)
- Master Machine Learning with Python — A deep dive into doing machine learning with scikit-learn constantly updated to showcase the latest and greatest tools. (300+ pages)

